Tags
animals, backbone, biology, Developmental biology, Gene, Gene expression, notochord, Popular science, science
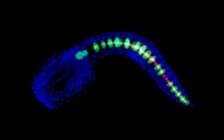 Genes have to be carefully coordinated to switch on at just the right moment in development in order to make a mature, complex embryo out of just a single cell. Scientists working at the Weill Medical College of Cornell University in New York have discovered how this coordination is accomplished. In a paper just published in PLOS Biology, they describe how the gene Brachyury controls the timing of a cascade of genes involved in a crucial process in vertebrate development.
Genes have to be carefully coordinated to switch on at just the right moment in development in order to make a mature, complex embryo out of just a single cell. Scientists working at the Weill Medical College of Cornell University in New York have discovered how this coordination is accomplished. In a paper just published in PLOS Biology, they describe how the gene Brachyury controls the timing of a cascade of genes involved in a crucial process in vertebrate development.
Brachyury is essential for the development of the notochord, a flexible rod which was the evolutionary precursor to the backbone. In vertebrates, the notochord is a important organizer in the early embryo but is later replaced by the backbone. Brachyury isn’t directly active in notochord development itself; instead, it’s a transcription factor, a gene that controls when and how other genes are expressed by binding to short stretches of DNA next to them. Animals with a defective copy of Brachyury can’t properly regulate the various genes involved in notochord development; without a notochord, the embryo dies.
In order to understand how Brachyury controls when genes are expressed, the team examined several of its target genes in the sea squirt Ciona intestinalis. They selected genes that were switched on at different times in notochord development and compared the Brachyury binding sites in them. It turned out that the genes activated earliest by Brachyury had several copies of the binding site, while genes which came on later had only one binding site. There was also a third class of genes which had no Brachyury binding sites; these genes, which were switched on latest, are indirectly activated by Brachyury via other transcription factors. The researchers hypothesized that the number of binding sites controlled when a gene got switched on, with fewer sites leading to later activation. To test this, they made a version of an early-onset gene in which some of the binding sites had been mutated so Brachyury could no longer bind to them; the mutated gene, with a reduced number of binding sites, was expressed later than the original version. “The full onset of its activity is delayed by a few hours, which is a crucial interval during the development of an organism,” said Dr. Yutaka Nibu, one of the authors. “Such a delay could cause a birth defect by slowing down the synthesis of a building block necessary for the organism to develop properly.”
Although this study showed how Brachyury controls when genes are expressed it notochord development, we still don’t know why the number of sites affects the timing. One possibility is that the presence of more binding sites might make it more likely that Brachyury will find one and bind to it; genes with more binding sites would therefore be sensitive to lower concentrations of the transcription factor. As Brachyury accumulates over time, it would be able to activate the less sensitive genes that have fewer binding sites. This sort of elegant simplicity is one of the reasons I’m endlessly fascinated by development. There may be grandeur in evolution, but there’s certainly beauty in development, that intricate choreography of interacting processes which builds complex structures while staying simple enough to be robust.
Refs
Lavanya Katikala, Hitoshi Aihara, Yale J. Passamaneck, Stefan Gazdoiu, Diana S. José-Edwards, Jamie E. Kugler, Izumi Oda-Ishii, Janice H. Imai, Yutaka Nibu, & Anna Di Gregorio (2013). Functional Brachyury Binding Sites Establish a Temporal Read-out of Gene Expression in the Ciona Notochord PLOS Biology, 11 (10) : 10.1371/journal.pbio.1001697
Lionel Christiaen (2013). Cis-Regulatory Timers for Developmental Gene Expression PLOS Biology, 11 (10) DOI: 10.1371/journal.pbio.1001698.g001

You must be logged in to post a comment.